Automated Ligand Searcher (ALISE)
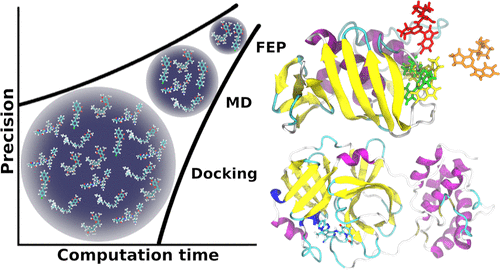
The Automated Ligand Searcher (ALISE) is designed as an automated computational drug discovery tool. To approximate the binding free energy of ligands to a receptor, ALISE includes a three-stage workflow, with each stage involving an increasingly sophisticated computational method: molecular docking, molecular dynamics, and free energy perturbation, respectively. The estimated binding free energy of a ligand is referred to as its score, and separate molecular docking, MD, and FEP scores are obtained in their respective stages. At the end of each stage, only the ligands with the best scores are selected to proceed to the next stage. This gradual segregation of ligands in each stage ensures that computational resources are not wasted by applying more computationally expensive methods to study poor drug candidates.
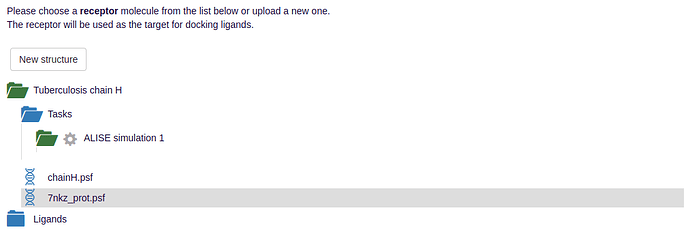
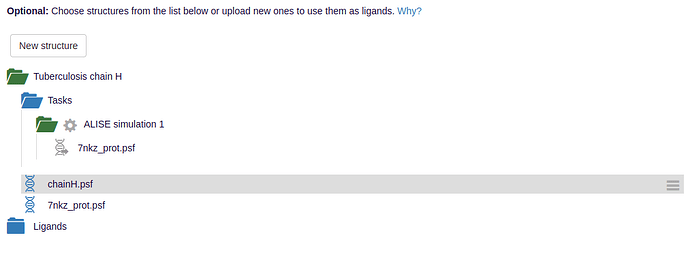
- To initiate the ALISE task, a receptor structure is required. This may be your typical pdb file which can optionally include a psf. ALISE has an automated psfgen-protocol, which can be skipped by uploading your own psf for the receptor.
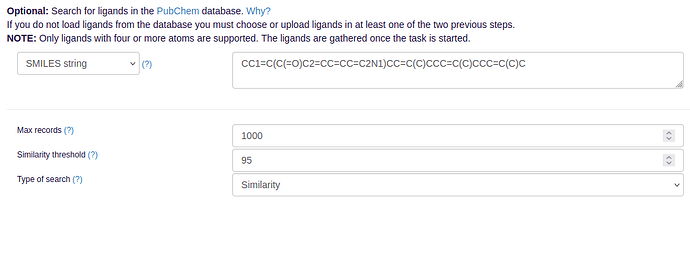
- You can then upload potential ligands either in MOL2 file format or again in pdb/psf. In addition, you can also initiate a ligand similarity search on PubChem by providing a substance’s SMILES string or molecular formula.
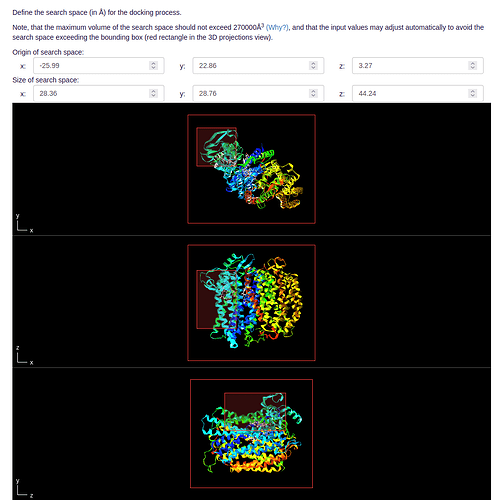
- After defining how many of your ligands should be considered in the MD and FEP simulations, you need to define a search space for Molecular Docking. This can be done automatically, but we suggest manually adjusting the area around your potential binding site:
- You can adjust the simulation parameters in the overview. Remember, as ALISE uses MD simulations, you may need to define your own potential by providing simulation parameters, if your receptor includes structures which are not included in the usual CHARMM parameter sets.
- ALISE will then run Molecular Docking, MD and FEP simulations and uses the best candidates of the previous step in the next simulation.
Example output
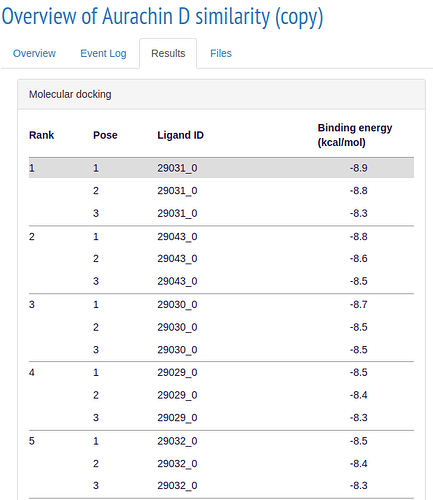
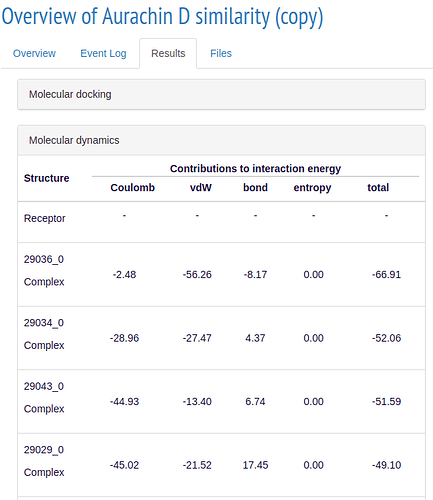
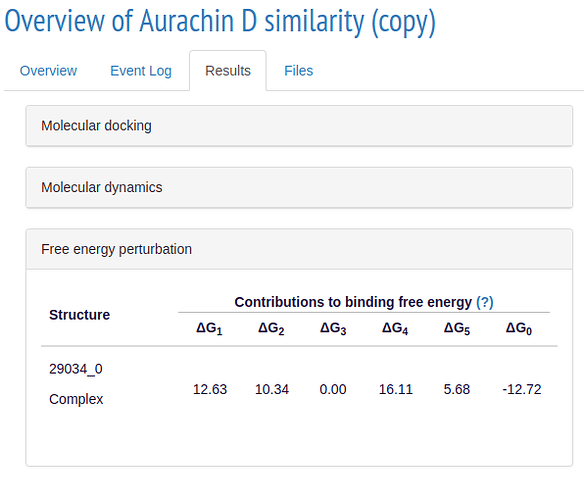
If your simulations ran succesfully, you will get results similar to these:
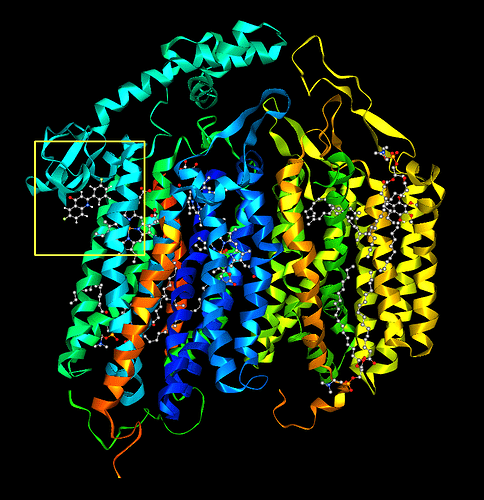
ALISE calculates a binding score for each ligand in the respective simulation step and ranks them accordingly. In the 3D model you can see how the ligand is located with respect to the receptor.
If you would like to gain a deeper understanding of how ALISE works we kindly invite you to read our corresponding paper Introducing the Automated Ligand Searcher.